Early prognosis of prostate most cancers (PCa) because the second most typical most cancers in males will not be related to exact and particular outcomes. Thus, alternate strategies with excessive specificity and sensitivity are wanted for correct and well timed detection of PCa. MicroRNAs regulate the molecular pathways concerned in most cancers by concentrating on a number of genes.
The aberrant expression of the microRNAs has been reported in completely different most cancers varieties together with PCa.
In this bioinformatics examine, we studied differential expression profiles of microRNAs and their goal genes in 4 PCa gene expression omnibus (GEO) databases.
PCa diagnostic biomarker candidates have been investigated utilizing bioinformatics instruments for evaluation of gene expression knowledge, microRNA goal prediction, pathway and GO annotation, in addition to ROC curves.
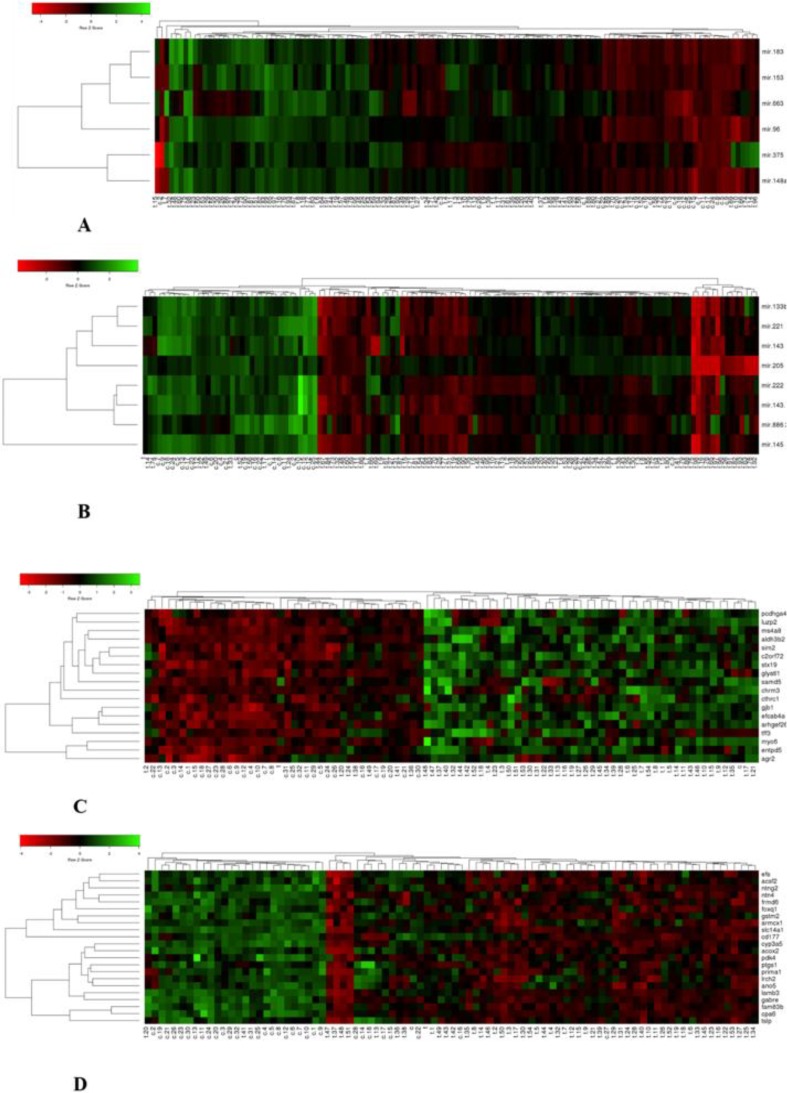
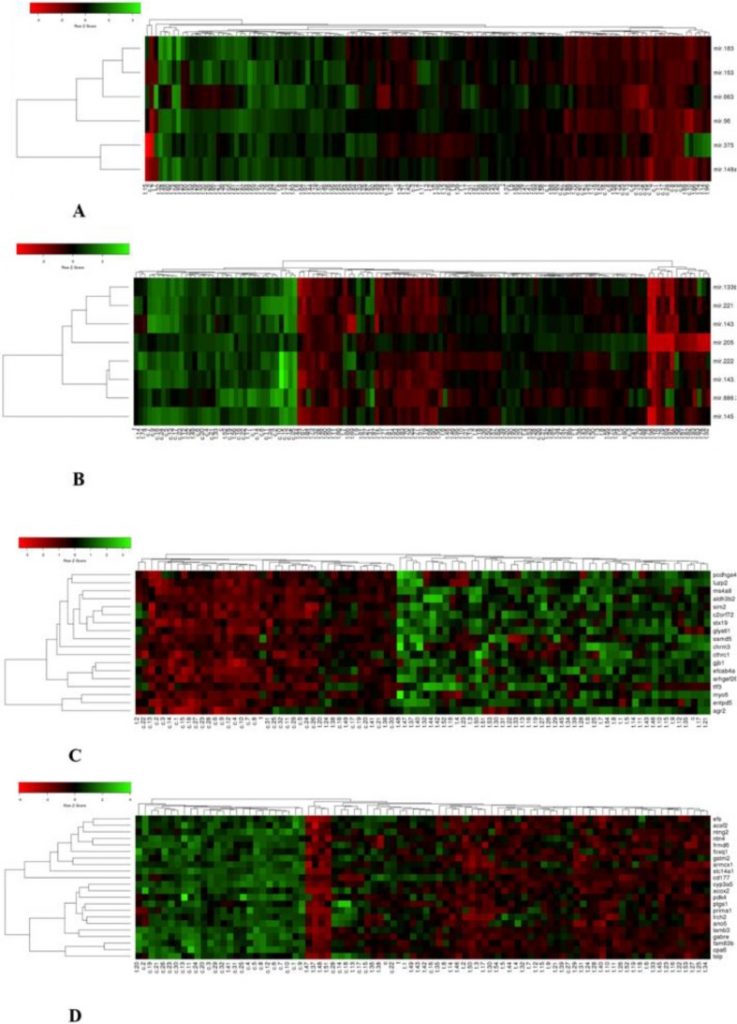
The outcomes of this examine revealed important modifications in the expression of 14 microRNAs and 40 related goal genes, which finally composed 4 mixture panels (miR- 375+96+663/ miR- 133b+143- 3p + 205/ C2ORF72 + ENTPD5 + GLYAT11/LAMB3 + NTNG2+TSLP) as candidate biomarkers succesful to differentiate between PCa tumor samples and regular prostate tissue samples. These biomarkers could also be recommended for a extra correct early prognosis of PCa sufferers together with present diagnostic exams.
Gene Expression Microarray Data Meta-Analysis Identifies Candidate Genes and Molecular Mechanism Associated with Clear Cell Renal Cell Carcinoma.
We aimed to discover potential molecular mechanisms of clear cell renal cell carcinoma (ccRCC) and present candidate goal genes for ccRCC gene remedy.
This is a bioinformatics-based examine. Microarray datasets of GSE6344, GSE781 and GSE53000 have been downloaded from Gene Expression Omnibus database. Using meta-analysis, differentially expressed genes (DEGs) have been recognized between ccRCC and regular samples, adopted by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) perform analyses. Then, protein-protein interplay (PPI) networks and modules have been investigated. Furthermore, miRNAs-target gene regulatory community was constructed.
ResultsTotal of 511 up-regulated and 444 down-regulated DEGs have been decided in the current gene expression microarray knowledge meta-analysis. These DEGs have been enriched in features like immune system course of and pathways like Toll-like receptor signaling pathway. PPI community and eight modules have been additional constructed.
A complete of 10 excellent DEGs together with TYRO protein tyrosine kinase binding protein (TYROBP), interferon regulatory issue 7 (IRF7) and PPARG co-activator 1 alpha (PPARGC1A) have been detected in PPI community. Furthermore, the miRNAs-target gene regulation analyses confirmed that miR-412 and miR-199b respectively focused IRF7 and PPARGC1A to manage the immune response in ccRCC.
ConclusionTYROBP, IRF7 and PPARGC1A may play necessary roles in ccRCC by way of participating in the immune system course of.